IC patients, especially those struggling with recurring flares, often wonder if they could have a chronic bacterial or fungal infection that is not being detected through traditional urine testing. A new, affordable and far more sensitive urine test, Next Generation DNA Sequencing, is now available that can identify both bacteria and fungi in a patients urine. This test is assisting physicians, infectious disease specialists and researchers in the diagnosis and treatment of complex microbial infections in the urinary tract. It is truly game changing!
CURIOUS? LEARN MORE HERE!
My Experience With Next Gen Testing
Next Generation DNA urine testing was brought to my attention last year by an IC patient who discovered that he had a hospital acquired drug resistant bladder infection. Traditional urine testing did not find the bacteria but Next Generation testing did and with proper treatment, that infection resolved. Interestingly, he had surgery earlier this year and again picked up another nosocomial infection in the hospital. And, again, Next Generation Sequencing not only identified the infection but also gave treatment suggestions that eliminated his infection. I was incredibly encouraged and very happy for him.
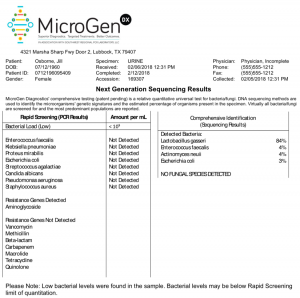
I decided to have my urine tested too because I had noticed that my urine was frequently cloudy after my hysterectomy last year. I was curious to see if I had any low grade infections. The test was very easy to do. They will mail you a kit and you then return a very small sample of your urine. My test results were fascinating. They found mostly beneficial bacteria however there was a very small population of three pathogenic bacteria: ecoli, actinomyces and enterococcus.
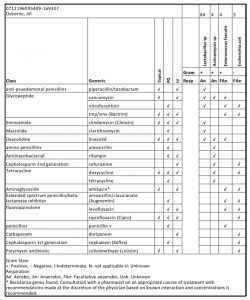
The question then became, should we treat this with antibiotics? MicroGen provides extensive drug resistance information in their test results as well. You can see my results attached below. They compare the DNA they find with known drug resistance data to create a master list of the most effective bacteria for your potential infection. Unlike the IC patient above, I chose not to take antibiotics because I was having no other symptoms of a UTI. I didn’t want to disrupt my gut biome with antibiotics when I wasn’t struggling with frequency, urgency or pain.
I am so compelled by the research that I am seeing with this new testing that I asked the company if we could be an affiliate and refer patients for the test. They said yes. Here’s an interview that I did with Rick Martin, the CEO of MicroGen Diagnostics. He explains the technology, how the testing is done and how patients can work with their physicians in understanding the results. If you’ve truly wondered if you had a chronic UTI and/or are struggling with candida in your urine, this test may answer that question once and for all.
An Interview with Rick Martin, CEO of MicroGen Diagnostics
- Why is the older, Agar plate culture being replaced with genomic testing?
- Is genomic testing more accurate than broth cultures?
- When in genomic testing used?
- How is genomic testing done?
- How accurate is genomic testing?
- How do you differentiate between good vs. bad bacteria?
- Will the test identify candida or other complex fungal infections?
- Will the test identify viral infections
- How does the test identify drug resistances?
- Does a doctor have to order the test? Is a physician signature required?
- How should a patient approach their doctor about having this test?
- What do the test results look like?
- Do you have any research data showing effectiveness of Microgen’s testing methods?
- Does Microgen provide treatment recommendations?
- How much does it cost? (Just $199)
- What to do if your doctor doesn’t believe the test results?
- Can a patient have another doctor order/review the test?
#1 – Why is the older, Agar plate culture being replaced with genomic testing?
Rick Martin: Agar plate was developed in 1870 by the father of microbiology, Robert Koch. This diagnostic form was developed to identify what Koch identified as pro- fessional pathogens. At the time, infections of typhoid, dysentery and so forth, and this technology relied on having to grow the microorgan- ism. We now know that less than 1% of all microorganisms can be grown in any type of medium so it’s much more accurate in terms of detecting microorganisms by identifying them by their unique DNA.
#2 – Some IC patients turned to broth culture testing with the intention of finding small amounts of fastidious infection. Is genomic testing more accurate than broth cultures?
Rick Martin: Culture technique is still a viable option for diagnosing acute infections. Acute infections are often caused by a single species or pathogen. They are not at all able to detect multiple species which form in biofilms. We now know that chronic infections are not caused by a single species but by multiple species living in these collaborative communities and attaching to the bladder wall, and the urinary tract.
They are highly synergistic, they have built a matrix around themselves and they are much more diffi- cult to detect because once the bacteria move into a biofilm phenotype, they express different genes, they behave entirely different, their metabolism is different and they don’t easily grow on any type of medium. If you are able to pull them from their cozy communities and drop them on a plate, they won’t grow, but if you did the same and got them to a lab where the DNA can be extracted, they can be identified by their DNA.
Culture, again, can only identify 1% of all microorganisms. In urinary samples alone, we’ve detected more than 2,000 species of microorgan- isms, both bacteria and fungus. In many cases, the samples are polymi- crobial, multiple species operating in a biofilm.
3. When is genomic testing used?
Rick Martin: Most practitioners are going to use it when they’ve already done a culture, they’ve already had a patient on antibiotics and the patient is still symptomatic. We would argue that it makes more sense to start using DNA testing versus using cultures again.
4. How is genomic testing done?
Rick Martin: It’s essentially no different than how you do a culture. Both diagnostic techniques require a urine sample. We need approxi- mately 8 ml of urine. We recommend that the urine is collected at different time points of the day, so early morning, midday, and end-of-day. You could put all three into one urine cup and then that would be transferred to an 8 ml vial for shipping and then put into a FedEx box and overnighted to our lab in Lubbock, Texas.
We also want to take into consid- eration things that will inhibit molecular testing, or what we call PCR inhibitors. Antibiotics have what we call active metabolites that accumalate in urine. If they are within the 8 ml vial of urine that you’re shipping they can degrade the DNA. There’s another product that patients are often treated with, Methenamine. This makes the urine more acidic, so it’s a PH issue. By causing it to be very acidic, it can degrade DNA during the shipping process. The best sample for molecular testing is one where you can stop all supplements, all antibiotics for at least a two-day period before collect- ing your urine and shipping it in.
We then perform two types of diagnostic tests. The first is a PCR, polymerase chain reaction, or rapid stream test, and that is followed by a full comprehensive next-generation sequencing.
5. How specific/accurate is genomic testing?
Rick Martin: We’re essentially 99% accurate.
6. How do you differentiate between good vs. bad, pathogenic bacteria?
Rick Martin: The greatest challenge for physicians in transferring to next generation DNA testing as a diagnostic tool is the interpretation. We now know that all urine has microorganisms and that the “micro- biome” within the urinary tract is going to vary by patient. It’s very difficult to say what a quote, “normal microbiome” is.
Essentially, when we test urine it is going to give you the current microbiome of that patient. You cannot use our test in isolation because if we find urine microorgan- isms in even healthy people that are asymptomatic, it doesn’t necessary mean you’re going to be treating those patients. The key point is you have to combine our test with other markers of infection or inflammation. If all other indications, whether it’s patients’ symptoms, burning urgency, whether it’s white blood cells count or leukocyte esterase assessment, whatever said rate CRP, whatever test you’re using, if all markers and symptoms point to infection, we are going to tell you with precision exactly what species are at the site. The clinician then has the decision to determine whether they wanted to go after the dominant species or they want to go after multiple species.
Some bacteria (i.e. Bacillus) could be considered normal flora. They could come from the vaginal cavity from women. We also know that in certain cases that Lactobacillus can also play a pathogenic role. Everything has to be looked at from host conditions and other values or indication of whether you’re fighting infection or not. It does make it a challenge. Given that all other values or diag- nostic test indicate infection and our test tells you what is found in the urine, then treat the dominant species and see how the patient responds.
If it’s a chronic infection you may have to use a localized delivery of antimicrobials to achieve the full killing power and to break through biofilms that form in the urinary tract from the bladder wall. In that case, you can add multiple antimi- crobials, antifungals, antibiotics into an irrigation and deliver them with other ingredients that break apart biofilms, lactoferrin, xylitol, EDTA and so forth and apply them in a localize delivery form which essen- tially gives you 5,000 times the killing power.
7. Will the test identify candida and/ or other complex fungal infections?
Rick Martin: Yes, we can identify all fungal species. Candida albicans is in our rapid screen PCR panel, but when we do full comprehensive, we will pick up every known mold and yeast. I believe we’ve identified more than 500 different fungal species in urine samples.
8. Will the test also identify potential viral infections? Have you seen/ tested viral infections?
Rick Martin: If you order the regular PCR and Next Generation Sequencing test, it will not detect viruses. Viruses are picked up using PCR technology. We do offer FDA approved panels that have viral primers which can detect different viruses, but the full comprehensive NGS is not designed to pick up viruses.
9. How does the test identify drug resistances?
Rick Martin: Bacteria develop resistance because they acquire a gene which basically turns off that antibiotic. Those genes now can be detected using PCR technology. We’ll build a primer for a specific gene, whether it’s the gene that causes beta-lactam resistance, which is basically cephalosporins and penicillin class, or quinolone resistance or aminoglycoside resistance. Each of these antibiotic classes has a known gene which causes resistance. We can detect those genes in the samples and tell the clinician whether you have, within your urine sample of resistance, two specific classes of antibiotics. We are going to tell you with precision exactly what species are at the site.
10. Does a doctor have to order the test? Does it require a physician signature?
Rick Martin: Certain states do allow for us to take samples from patients without a physician’s signa- ture, other states require a physician signature. We provide a list of those states that do not require a physi- cian’s signature on our website. If you are in one of those states that does not require a physician’s signature, you can take and follow the directions for a urine sample and send it directly to us.
11. If a patient wants to have the test done but the doctor minimizes the results or, worse, suggests agar plate testing, how should the patient approach their doctor? Do you have supporting documentation that he/she can share with their clinician?
Rick Martin: Yes, we do. There are a number of papers to show that cultures can only detect 1% of all microorganisms and there are a number of papers to show that sequencing is more accurate. It doesn’t take that much in terms of understanding that cultures have to grow the microbes, DNA or molecular diagnostics relying on extraction of the DNA, identify the species by its DNA.
We also have done head to head clinical trials to show that if an antibiotic decision is made on culture versus DNA, that when the antibiotic decision is based on DNA, it’s more accurate because patients’ symptoms came down faster, and more of them cleared up their infec- tion than with culture.
If a patient has already had culture, and already had antibiotics and they should simply say “Well, we’ve already done the same thing, we already tried cultures, why are we doing the same thing again? Why don’t we try a new diagnostic technique? Because I’m still symptomatic and still suffering.”
12. What do the test results look like? (i.e. quick PCR results vs. 5-day genomic results)
Rick Martin: PCR is delivered overnight, so if you take your urine sample on Sunday and shipped it to us on Monday, by Tuesday afternoon you’ll have the rapid screen PCR which is 21 species plus resistant genes. Then two days later, you’ll have your full comprehensive next gen sequencing. The delivery time for the full comprehensive NGS is right around 3 to 4 days.
13. Do you have any research data showing effectiveness of Microgen’s testing methods?
Rick Martin: When it comes to using molecular as a diagnostic tool for detection of microorganisms, we have more published clinical trials than any lab in the world. We have published studies in urinary tract infections, where we were statistically significantly superior to culture in relieving patients’ symptoms scores and clearing infections from UTIs.
We also have published studies in prosthetic joint infections, where we have shown superiority over culture in terms of detecting the microbes or polymicrobial infections that cause infections in infected knees, and infected hips.
We have four ongoing studies that are looking at patients suffering from chronic sinus infections. We’ve had multiple studies published in wound care that are looking at sequencing versus culture, improving sequencing is superior to detecting and treating most of the biofilms or multiple species to polymicrobial infections that cause delayed healing in wounds.
14. Given the deep concern with antibiotic resistance, does Microgen provide therapeutic recommendations for treatment that also reflect concerns about over treatment?
Rick Martin: Using our technology, you’re able to know with preci- sion exactly which species is domi- nant in the sample and you’re able to make your antibiotic selection based on solid DNA evidence. Our test is not forced, there’s nothing that says you have to treat based on what we’ve detected. Again, you’re going to combine our diagnostic informa- tion with the other information you have to indicate whether there’s an infection or not. Then you can make the decision as to which species you want to treat.
You’re better off knowing the full and complete picture of what’s there at the site versus not knowing and guessing and going empirically or broad spectrum hoping that you have picked the right combination of antibiotics to get a good clinical outcome. If anything, our test is going to improve antibiotic stewardship by helping physicians select the right antibiotic for the right patient, right bug.
15. How much does genomic testing cost?
Rick Martin: The barrier to molecular testing has been time and cost. When the NGS was first avail- able it would cost thousands of dollars to run one sample, and it would take weeks to a month to deliver the results. We now have it down to an average of a 3 to 4 day turnaround time and if their insurance doesn’t cover it, we do it for less than $200.
If you go to a lab, aerobic cultures run around $70. Anaerobic, which is a separate test, because again when you’re doing culture you’re having to pay for different medias and different types of bacteria, aerobic, anaerobic and then obviously you have fungus. An anaerobic test usually runs about $160. Then fungus, which is more difficult and time consuming, I think it can take a couple of weeks, and it runs anywhere between $500 to $600.
With our technology you’re not growing different plates for aerobic, anaerobic and fungus. If those species are in the sample we will extract the DNA and detect them and identify them by their DNA all in one test for $200. It becomes both more accurate and less expensive than traditional culture.
16. The results are in, but my doctor doesn’t believe them. How should that patient approach their clinician?
Rick Martin: This is a common scenario. If the physician given the report doesn’t believe the results, we can show that we have proven concordance. We’ve done studies that show if a traditional urine culture grew Staph where we are also able to identify or detect the Staph 96.1% of the time using DNA testing.
You can say, “Well doc, you treated based on culture and my culture in the past has either been negative or “what- ever it was” grew. You treated it and I didn’t get better. This test is saying that my urine has these types of bacteria and/or fungi. I’ve never been treated for that. I would like you to now treat me for the dominant species based on DNA evidence versus culture. I have every- thing to gain and nothing to lose by doing this. I would really like you to be open-minded to making a treatment decision based on this evidence versus doing another culture again.”
17. If a urologist sees no value in additional testing, can a patient ask another doctor to order/review the test?
Rick Martin: Yes, and we can provide help on that. We have infectious disease doctors who will do a phone consult with the patient and walk them through the results. We can also have the infectious disease doctor notify their doctor and talk to their doctor about how the test has been utilized and how beneficial it has been.